


| Dynamic Programming |
Programming Assignment |
Standard Libraries. In order to complete this assignment, you may need copies of StdIn.java and Picture.java in the same directory as your program.
Because this assignment has more parts than most, it will be worth 30 points.
| Background: Interfaces |
Objects gives us a way to specify an API for a custom data type through which clients can access and manipulate the data. The API gives us a well-defined series of operations and a contract as to what result to expect. It does not say how that result is achieved, and we are free to implement it any way we want, or the change it.
The limitation of the simple objects we have seen so far is that we can have only one implementation of the same behavior at a time. But what if we want to have two very different implementations at the same time? For instance, it is meaningful to define an operation "compare two things," but the way we implement that comparison will be very different for strings and integers. With a mechanism to define the operation "compare two things," that we can use for both strings and integers, we could write a single sort method that sorts strings alphabetically and integers numerically. In other words, we'd like a way to express that multiple different class implement the same API.
Java's mechanism for expressing that multiple classes implement the same API is called interfaces. An interface looks very similar to a class, but it contains no instance variables, and only method signatures without implementations:
The Comparable interface is part of the standard library and provides an API that any class can implement so that clients can compare it. Object is a special: Java will convert any object type (String, RingBuffer, etc.) to Object automatically, and you can explicitly convert back to the original type. It comes up in many interface specifications, but you won't need it for this assignment or for the course.
public interface Comparable { public int compareTo(Object o); }
A sort method can rely this interface to know how to sort any object that respects the Comparable API:
public class SelectionSort { public static void sort(Comparable[] arr) { for (int i = 0; i < arr.size(); i++) { for (int j = i + 1; } }
Note that SelectionSort can only use the methods announced in the Comparable interface, but in exchange it automatically works with every class that implements it.
When you write a class, you can elect to implement the Comparable interface, which means you include all the methods specified in the interface in your class. SelectionSort will "just work" with your class
public class MyInteger implements Comparable { public int value; public int compareTo(Object o) { MyInteger mi = (MyInteger) o; if (value < mi.value) return -1; else if (value > mi.value) return 1; else return 0; } }
The statement implements Comparable is the contract with Java that your class will include all the methods from the Comparable interface. If it doesn't, your class will not compile. That contract in turn allows classes like SelectionSort to work with any method that implements Comparable since it knows the methods it requires will exist.
In this assignment you will compute DNA sequence alignments and also the best "seam" along which to stitch two images together. The optimization process involves evaluating how well individual base pairs match up in the DNA sequence case, and how similar overlapping image pixels are in the image stitching case. Although these two things are completely different, the algorithm to find a good solution based on those "costs" turns out to be the same. You will therefore write a single class to find an optimal solution for both problems. For each one, you will write a class that implements a interface to calculate the costs on which the optimization relies.
| Part 1: Global Sequence Alignment |
Write a program to compute the optimal sequence alignment of two DNA strings. This program will introduce you to the emerging field of computational biology in which computers are used to do research on biological systems. Further, you will be introduced to a powerful algorithmic design paradigm known as dynamic programming.
Biology review. A genetic sequence is a string formed from a four-letter alphabet {Adenine (A), Thymine (T), Guanine (G), Cytosine (C)} of biological macromolecules referred to together as the DNA bases. A gene is a genetic sequence that contains the information needed to construct a protein. All of your genes taken together are referred to as the human genome, a blueprint for the parts needed to construct the proteins that form your cells. Each new cell produced by your body receives a copy of the genome. This copying process, as well as natural wear and tear, introduces a small number of changes into the sequences of many genes. Among the most common changes are the substitution of one base for another and the deletion of a substring of bases; such changes are generally referred to as point mutations. As a result of these point mutations, the same gene sequenced from closely related organisms will have slight differences.
The problem. Through your research you have found the following sequence of a gene in a previously unstudied organism.
A A C A G T T A C C
What is the function of the protein that this gene encodes? You could begin a series of uninformed experiments in the lab to determine what role this gene plays. However, there is a good chance that it is a variant of a known gene in a previously studied organism. Since biologists and computer scientists have laboriously determined (and published) the genetic sequence of many organisms (including humans), you would like to leverage this information to your advantage. We'll compare the above genetic sequence with one which has already been sequenced and whose function is well understood.
If the two genetic sequences are similar enough, we might expect them to have similar functions. We would like a way to quantify "similar enough."T A A G G T C A
Edit-distance. In this assignment we will measure the similarity of two genetic sequences by their edit distance, a concept first introduced in the context of coding theory, but which is now widely used in spell checking, speech recognition, plagiarism detection, file revisioning, and computational linguistics. We align the two sequences, but we are permitted to insert gaps in either sequence (e.g., to make them have the same length). We pay a penalty for each gap that we insert and also for each pair of characters that mismatch in the final alignment. Intuitively, these penalties model the relative likeliness of point mutations arising from deletion/insertion and substitution. We produce a numerical score according to the following table, which is widely used in biological applications:
operation cost insert a gap 2 align two characters that mismatch 1 align two characters that match 0
Here are two possible alignments of the strings x = "AACAGTTACC" and y = "TAAGGTCA":
x y cost ------------ A T 1 A A 0 C A 1 A G 1 G G 0 T T 0 T C 1 A A 0 C - 2 C - 2 --- 8x y cost ------------ A T 1 A A 0 C - 2 A A 0 G G 0 T G 1 T T 0 A - 2 C C 0 C A 1 --- 7
The first alignment has a score of 8, while the second one has a score of 7. The edit-distance is the score of the best possible alignment between the two genetic sequences over all possible alignments. In this example, the second alignment is in fact optimal, so the edit-distance between the two strings is 7. Computing the edit-distance is a nontrivial computational problem because we must find the best alignment among exponentially many possibilities. For example, if both strings are 100 characters long, then there are more than 10^75 possible alignments.
We will explain a recursive solution which is an elegant approach. However it is far too inefficient because it recalculates each subproblem over and over. Once we have defined the recursive definition we can redefine the solution using a dynamic programming approach which calculates each subproblem once.
A recursive solution. We will calculate the edit-distance between the two original strings x and y by solving many edit-distance problems on the suffixes of the two strings. We use the notation x[i] to refer to character i of the string. We also use the notation x[i..M] to refer to the suffix of x consisting of the characters x[i], x[i+1], ..., x[M-1]. Finally, we use the notation opt[i][j] to denote the edit distance of x[i..M] and y[j..N]. For example, consider the two strings x = "AACAGTTACC" and y = "TAAGGTCA" of length M = 10 and N = 8, respectively. Then, x[2] is 'C', x[2..M] is "CAGTTACC", and y[8..N] is the empty string. The edit distance of x and y is opt[0][0].
Now we describe a recursive scheme for computing the edit distance of x[i..M] and y[j..N]. Consider the first pair of characters in an optimal alignment of x[i..M] with y[j..N]. There are three possibilities:
opt[i][j] = min { opt[i+1][j+1] + 0/1, opt[i+1][j] + 2, opt[i][j+1] + 2 }
| 0 1 2 3 4 5 6 7 8
x\y | T A A G G T C A -
-----------------------------------
0 A | 7 8 10 12 13 15 16 18 20
1 A | 6 6 8 10 11 13 14 16 18
2 C | 6 5 6 8 9 11 12 14 16
3 A | 7 5 4 6 7 9 11 12 14
4 G | 9 7 5 4 5 7 9 10 12
5 T | 8 8 6 4 4 5 7 8 10
6 T | 9 8 7 5 3 3 5 6 8
7 A | 11 9 7 6 4 2 3 4 6
8 C | 13 11 9 7 5 3 1 3 4
9 C | 14 12 10 8 6 4 2 1 2
10 - | 16 14 12 10 8 6 4 2 0
A dynamic programming approach. A direct implementation of the above recursive scheme will work, but it is spectacularly inefficient. If both input strings have N characters, then the number of recursive calls will exceed 2^N. To overcome this performance bug, we use dynamic programming. (Read the first section of Section 9.6 for an introduction to this technique.) Dynamic programming is a powerful algorithmic paradigm, first introduced by Bellman in the context of operations research, and then applied to the alignment of biological sequences by Needleman and Wunsch. Dynamic programming now plays the leading role in many computational problems, including control theory, financial engineering, and bioinformatics, including BLAST (the sequence alignment program almost universally used by molecular biologist in their experimental work). The key idea of dynamic programming is to break up a large computational problem into smaller subproblems, store the answers to those smaller subproblems, and, eventually, use the stored answers to solve the original problem. This avoids recomputing the same quantity over and over again. Instead of using recursion, use a nested loop that calculates opt[i][j] in the right order so that opt[i+1][j+1], opt[i+1][j], and opt[i][j+1] are all computed before we try to compute opt[i][j].
Recovering the alignment itself. The above procedure describes how to compute the edit distance between two strings. We now outline how to recover the optimal alignment itself. The key idea is to retrace the steps of the dynamic programming algorithm backwards, re-discovering the path of choices (highlighted in red in the table above) from opt[0][0] to opt[M][N]. To determine the choice that led to opt[i][j], we consider the three possibilities:
x y cost ------------ A T 1 A A 0 C - 2 A A 0 G G 0 T G 1 T T 0 A - 2 C C 0 C A 1
API specification. Your program should consist of a class Match.java with one static method to compute the optimal match between a pair of objects. It relies on the DataPair interface to compute the costs so that it can work with different types of objects. The optimal solution is returned as a linked list of Path nodes. Each Path node stores the row and column of the element it represents, the total cost of the match from that point to the end, and a pointer to the next path node. So for opt matrix above, the Path nodes would follow the red numbers: the row and col variables of the first node would both be 0 and cost would be 7. In the second node, row and col would be 1 and cost would be 6, and so on.
public class Match
--------------------------------------------------------------------------------
static Path match(DataPair dp) // return the optimal match between
// the items specified by dp
//
// return null if dp == null or
// if dp.rows() == 0 or dp.cols() == 0
static void main(String[] args) // method to test Match (see Checklist)
public interface DataPair
--------------------------------------------------------------------------------
public int rows(); // number of rows in the matrix
public int cols(); // number of columns in the matrix
public int cost(int row, int col, int dir);
// cost computes the cost of a path running
// through (col, row) assuming it is coming
// from the direction specified by dir
//
// if dir is -1 we are moving horizontally
// (i.e. from (col - 1, row) to (col, row)
// if dir is +1 we are moving vertically
// (i.e. from (col, row - 1) to (col, row)
// if dir is 0, we are moving diagonally
public int baseCost(int row, int col);
// baseCost computes the cost of a
// path running through (col, row)
// without considering the direction
// the path took to get there
public class Path -------------------------------------------------------------------------------- public int row, col; // the row and column this node represents public int cost; // the matching cost from this point on public Path next; // the next node in the optimal path
You must also write a class EditDistance.java that implements the DataPair interface for matching strings, and includes a main method that reads two strings from standard input, calls Match.match() to find their optimal alignment, and prints the edit distance, optimal alignment, and associated penalties to standard output.
public class EditDistance implements DataPair
--------------------------------------------------------------------------------
public int rows() // length of the first string + 1
// (i.e. number of rows in the
// alignment matrix)
public int cols() // length of the second string + 1
// (i.e. number of columns in the
// alignment matrix)
public int cost(int row, int col, int dir)
// The cost of matching at position
// (col, row) in the alignment matrix
// assuming we are following the direction
// specified by dir
public int baseCost(int row, int col)
// The cost of matching at position
// (col, row) without regard to direction
// For EditDistance this is the same
// as the cost assuming we are moving
// diagonally.
public EditDistance(String x, String y)
// create an EditDistance object
// for the strings x and y
static void main(String[] args) // read 2 strings from standard input.
// compute and print the edit distance
// between them and output an optimal
// alignment and associated penalties.
Your program. Write a program EditDistance.java that conforms to the API above and whose main method reads, from standard input, two strings of characters, creates an EditDistance object for them, and computes the optimal matching between them using Match.match(). (Although, in the application described, the characters represent genetic sequences, your program should handle any sequence of alphanumeric characters.) Your program should then print the edit distance between the two strings, and print out the optimal match along with the individual penalties using the following format:
% java EditDistance < example10.txt
Edit distance = 7
A T 1
A A 0
C - 2
A A 0
G G 0
T G 1
T T 0
A - 2
C C 0
C A 1
Testing The archive sequence.zip contains short test data files and actual genomic data files. In addition, you should create test cases of your own to make sure you handle all special cases correctly.
| Part 2: Image Stitching |
Write a program to stitch two images together into a larger, panoramic image. Combining multiple images into a larger panorama is one of the oldest techniques in the field of computational photography, which combines computer algorithms with traditional photography to create images that are difficult or impossible to achieve purely with optical lenses. Many recent cameras are capable of building panoramas out of individual pictures automatically. Surprisingly, a key part of the process involves the same kind of optimization as sequence alignment. The core if this program will therefore be the Match class that you wrote in Part 1. You will not need to modify this class at all.
The Problem. Camera lenses have a much narrower field of view than human vision, so a single photograph cannot capture an entire landscape as you see it. Panoramic stitching gives a solution to the problem by taking two overlapping images and "stitching" them together into a single, large image. The same technique can be used to insert part of one image into another, for instance yourself into a historic picture.
Unless the images overlap perfectly, we need to combine them together cleverly so they appear to be one single image. One of the most successful way to do this is to find a "seam" where the two image are very similar. Everything to the left of that seam is taken from the left-hand image, and everything from the right is taken from the right-hand image. As long as the two images are nearly identical along that seam, it won't be visible even if the images are very different on either side of it:
 |
 |
 |
| First input image | Second input image | Two images stitched along the black line |
 |
 |
 |
| First input image | Second input image | Two images stitched along the least visible seam |
Finding the optimal seam. Let us start with the simple case where the images overlap perfectly, and we would like to fine a seam that runs from the upper-left corner to the lower-right corner (as in the red and green images above). Every pixel in the output image that is on the seam or to the right of it should come from the first image. Every pixel that is to the left of the seam should come from the second image. We will also restrict the seam to move right, down, or diagonally right and down. The seam is not allowed to wiggle back and forth like an S.
Along an optimal seam, the pixel values in the two images will be identical. When we can't find such a seam, we would like the seam to pass through locations where the pixel values in the two images are as similar as possible. Recalling that a pixel color is a combination of red, green, and blue values, we define the squared pixel difference to be: D = (R1 - R2)2 + (G1 - G2)2 + (B1 - B2)2 . If we take the squared pixel difference to be the cost of letting the seam run through a particular location in the images, and the cost of a particular seam as the sum of squared differences of all pixels along the seam, we end up with the same dynamic programming problem as with sequence alignment, and we can solve it using Match.match(). Location (i, j) in the optimization matrix will track the cost of letting the seam run through pixel location (i, j) rather than the cost of matching character i in the first string to character j in the second string. But this difference in interpretation of the data has no effect on the process, so the same algorithm works for both problems.
Stitching two perfectly overlapping images is of limited value of course. The more common case where the images only partially overlap is shown in the sea image. We can reduce this to the simple case by ignoring the non-overlapping portions. The seam could run from upper-left to lower-right or from upper-right to lower-left depending on how the images overlap, but we can deal with this by flipping the images so that the seam always runs from upper-left to lower-right:
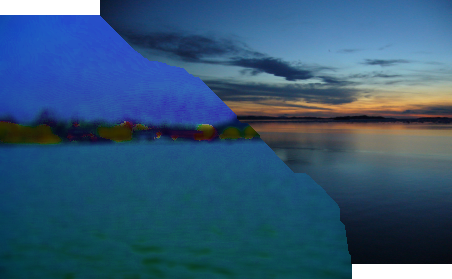 |
 |
 |
 |
We provide the program Stitcher.java to do this cropping and stitching for you. It finds the overlapping portions of the two images, rotates them so the seam runs from upper-left to lower-right, calls the ImageSeam class that you will write to stitch these together, then inserts the result into a combined panorama. The stitched sea images were create with the following command that specifies the two input images and number of pixels to shift the second image relative to the first image (positive numbers shift the second image right and down, negative numbers shift it left and up):
% java Stitcher sea1.png sea2.png 318 -7
Program and API Specification. Write a class ImageSeam to compute the optimal seam between two Pictures using Match.match(). Your class should compute the optimal path in the constructor, and store it and the two Pictures instance variables. Client programs can then call getStitched() to retrieve a new, stitched image. You class must conform to the following API:
public class ImageSeam implements DataPair
--------------------------------------------------------------------------------
public int rows(); // number of rows in the images
public int cols(); // number of columns in the images
public int cost(int row, int col, int dir);
// return the base cost of a seam running
// through (col, row); ignore dir because
// the direction the seam runs does not
// affect the total cost in this application
public int baseCost(int row, int col);
// return the squared pixel difference
// between the two images at (col, row)
public Picture getStitched(); // return a new image stitched along the
// optimal seam between a and b based
// on the path computed in the constructor
//
// if the optimal path is null, return null
public ImageSeam(Picture a, Picture b)
// compute the optimal path between a and b
// using Match.match()
//
// if a or b is null, or the width or height
// of a or b is zero, or if the width or
// height of a and b differ, set the optimal
// path to null
Because you will be working with Color objects, you should include the statement
import java.awt.Color;
at the top of your file, before the class declaration. You can test this program using Stitcher.java, but you can also write your own main() method in ImageSeam that runs simpler tests if you wish.
Testing. You can use the images from the stitched examples above (example-green.png, example-red.png, example-stitched.png, sea1.png, sea2.png, and sea-stitched.png to test you ImageSeam class. We generated the stitched version by running the following commands:
% java Stitcher example-green.png example-red.png 0 0 % java Stitcher sea1.png sea2.png 318 -7
Stitcher.java also contains a testing mode to help you test your ImageSeam. You can optionally specify a reference image as the last argument, and it will show where your stitched version differs from the reference images. For instance,
% java Stitcher sea1.png sea2.png 318 -7 sea-stitched.png
will display an all white image if your stitched version is identical to our "ground truth" image sea-stitched.png. Any pixels that are different will be colored black. Note that you may get some black pixels even if the program is correct because there are sometimes more than one seam that have the same total cost.
In addition to your ImageSeam.java you should submit a zip file containing at least two pairs of test images that you used to test your program (and the result of stitching them). You may use existing images or create your own. We will award one point of extra credit if at least one of your image pairs is particularly clever or creative. Include instructions in your readme.txt file for running Stitcher with your image pairs.
Checklist. Don't forget to read the checklist.
Submission. Submit the files Match.java, EditDistance.java, ImageSeam.java, a zip archive images.zip containing your images, and your readme.txt. Your images must be contained in a zip archive, not in any other format (e.g. rar), or you will receive a significant deduction for breaking our testing scripts.